import sklearn.datasets
from sklearn.ensemble import RandomForestRegressor
from model_inspector import get_inspectorAny Model
X_diabetes, y_diabetes = sklearn.datasets.load_diabetes(return_X_y=True, as_frame=True)
inspector = get_inspector(
RandomForestRegressor().fit(X_diabetes, y_diabetes), X_diabetes, y_diabetes
)/Users/greg.gandenberger/repos/model_inspector/model_inspector/inspect/any_model.py:56: UserWarning: `model` does not have the `feature_names_in_`
attribute, so we cannot confirm that `model`'s feature
names match `X`'s column names. Proceed at your own
risk!
warnings.warn(_Inspector.methods
_Inspector.methods ()
Show available methods.
inspector.methods['_check_cols',
'permutation_importance',
'plot_feature_clusters',
'plot_partial_dependence',
'plot_permutation_importance',
'plot_pred_vs_act',
'plot_residuals',
'show_correlation']_Inspector.permutation_importance
_Inspector.permutation_importance (sort:bool=True, scoring=None, n_repeats=5, n_jobs=None, random_state=None, sample_weight=None, max_samples=1.0)
Calculate permutation importance.
Parameters:
sort: Sort features by decreasing importance.
Remaining parameters are passed to sklearn.inspection._permutation_importance.permutation_importance.
inspector.permutation_importance()bmi 0.482974
s5 0.472974
bp 0.137960
s6 0.092853
s3 0.088472
age 0.077993
s2 0.067045
s1 0.051492
sex 0.023468
s4 0.022805
dtype: float64_Inspector.plot_partial_dependence
_Inspector.plot_partial_dependence (categorical_features=None, feature_names=None, target=None, response_method='auto', n_cols=3, grid_resolution=100, percentiles=(0.05, 0.95), method='auto', n_jobs=None, verbose=0, line_kw=None, ice_lines_kw=None, pd_line_kw=None, contour_kw=None, ax=None, kind='average', centered=False, subsample=1000, random_state=None)
Plot partial dependence.
Returns NumPy array of Axes objects.
Remaining parameters are passed to sklearn.inspection._plot.partial_dependence.from_estimator.
axes = inspector.plot_partial_dependence(features=["bp", "bmi", ["bp", "bmi"]])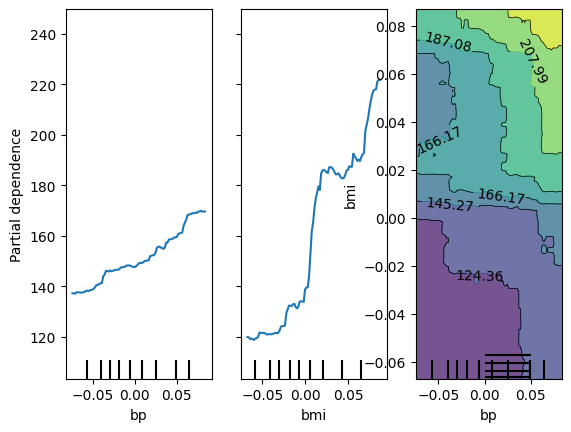
_Inspector.plot_feature_clusters
_Inspector.plot_feature_clusters (corr_method:str='spearman', ax:matplotlib.axes._axes.Axes=None, p=30, truncate_mode=None, color_threshold=None, get_leaves=True, orientation='top', labels=None, count_sort=False, distance_sort=False, show_leaf_counts=True, no_plot=False, no_labels=False, leaf_font_size=None, leaf_rotation=None, leaf_label_func=None, show_contracted=False, link_color_func=None, above_threshold_color='C0')
Plot a dendrogram based on feature correlations.
Parameters:
corr_method: Method of correlation to pass todf.corr()ax: MatplotlibAxesobject. Plot will be added to this object if provided; otherwise a newAxesobject will be generated.
Remaining parameters are passed to model_inspector.explore.plot_column_clusters.
ax = inspector.plot_feature_clusters()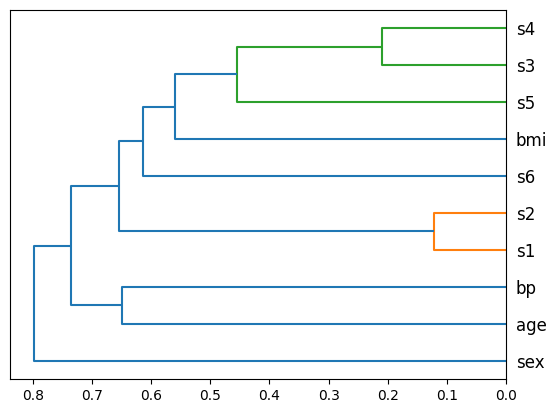
_Inspector.plot_permutation_importance
_Inspector.plot_permutation_importance (ax:Optional[matplotlib.axes._axe s.Axes]=None, importance_kwargs:O ptional[dict]=None, plot_kwargs:Optional[dict]=None)
Plot a correlation matrix for self.X and self.y.
Parameters:
ax: MatplotlibAxesobject. Plot will be added to this object if provided; otherwise a newAxesobject will be generated.importance_kwargs: kwargs to pass tosklearn.inspection.permutation_importanceplot_kwargs: kwargs to pass topd.Series.plot.barh
ax = inspector.plot_permutation_importance()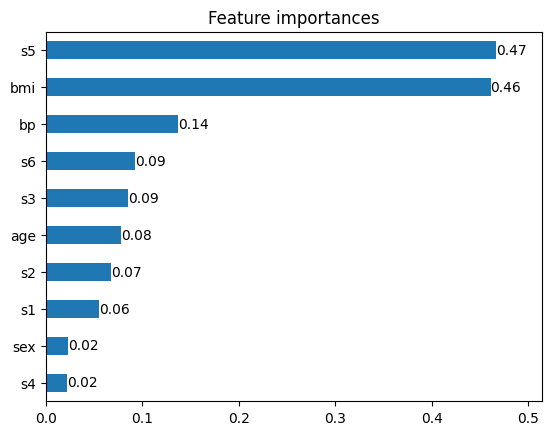
_Inspector.show_correlation
_Inspector.show_correlation (method='pearson', cmap:str|Colormap='PuBu', low:float=0, high:float=0, axis:Axis|None=0, subset:Subset|None=None, text_color_threshold:float=0.408, vmin:float|None=None, vmax:float|None=None, gmap:Sequence|None=None)
Show a correlation matrix for self.X and self.y.
If output is not rendering properly when you reopen a notebook, make sure the notebook is trusted.
Remaining parameters are passed to model_inspector.explore.show_correlation.
inspector.show_correlation()| age | sex | bmi | bp | s1 | s2 | s3 | s4 | s5 | s6 | target | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| age | 1.00 | 0.17 | 0.19 | 0.34 | 0.26 | 0.22 | -0.08 | 0.20 | 0.27 | 0.30 | 0.19 |
| sex | 0.17 | 1.00 | 0.09 | 0.24 | 0.04 | 0.14 | -0.38 | 0.33 | 0.15 | 0.21 | 0.04 |
| bmi | 0.19 | 0.09 | 1.00 | 0.40 | 0.25 | 0.26 | -0.37 | 0.41 | 0.45 | 0.39 | 0.59 |
| bp | 0.34 | 0.24 | 0.40 | 1.00 | 0.24 | 0.19 | -0.18 | 0.26 | 0.39 | 0.39 | 0.44 |
| s1 | 0.26 | 0.04 | 0.25 | 0.24 | 1.00 | 0.90 | 0.05 | 0.54 | 0.52 | 0.33 | 0.21 |
| s2 | 0.22 | 0.14 | 0.26 | 0.19 | 0.90 | 1.00 | -0.20 | 0.66 | 0.32 | 0.29 | 0.17 |
| s3 | -0.08 | -0.38 | -0.37 | -0.18 | 0.05 | -0.20 | 1.00 | -0.74 | -0.40 | -0.27 | -0.39 |
| s4 | 0.20 | 0.33 | 0.41 | 0.26 | 0.54 | 0.66 | -0.74 | 1.00 | 0.62 | 0.42 | 0.43 |
| s5 | 0.27 | 0.15 | 0.45 | 0.39 | 0.52 | 0.32 | -0.40 | 0.62 | 1.00 | 0.46 | 0.57 |
| s6 | 0.30 | 0.21 | 0.39 | 0.39 | 0.33 | 0.29 | -0.27 | 0.42 | 0.46 | 1.00 | 0.38 |
| target | 0.19 | 0.04 | 0.59 | 0.44 | 0.21 | 0.17 | -0.39 | 0.43 | 0.57 | 0.38 | 1.00 |